There’s a clear connection between the phenomenon of self-assembly, by which objects at the nanoscale arrange themselves into complex shapes by virtue of programmed patterns of stickiness, and information. The precisely determined three dimensional shape of a protein is entirely specified by the one-dimensional sequence of amino acids along the chain, and the information that specifies this sequence (and thus the shape of the protein) is stored as a sequence of bases on a piece of DNA. If one is talking about information, it’s natural to think of computing, so its natural to ask whether there is any general relationship between computing processes, thought of at their most abstract, and self-assembly.
The person who has, perhaps, done the most to establish this connection is Erik Winfree, at Caltech. Winfree’s colleague, Paul Rothemund, made headlines earlier this year by making a nanoscale smiley face, but I suspect that less well publicised work the pair of them did a couple of years ago will prove just as significant in the long run. In this work, they executed a physical realisation of a cellular automaton whose elements were tiles of DNA with particular patches of programmed stickiness. The work was reported in PLoS Biology here; see also this commentary by Chengde Mao. A simple one-dimensional cellular automaton consists of a row of cells, each of which can take one of two values. The automaton evolves in discrete steps, with a rule that determines the value of a cell on the next step by reference to the values of the adjacent cells on the previous step (for an introduction, to elementary cellular automata, see here). One interesting thing about cellular automata is that very simple rules can generate complex and interesting patterns. Many of these can be seen in Stephen Wolfram’s book, A New Kind of Science, (available on line here. It’s worth noting that some of the grander claims in this book are controversial, as is the respective allocation of credit between Wolfram and the rest of the world, but it remains an excellent overview of the richness of the subject).
I can see at least two aspects of this work that are significant. The first point follows from the fact that a cellular automaton represents a type of computer. It can be shown that some types of cellular automaton are, in fact, equivalent to universal Turing machines, able in principle to carry out any possible computation. Of course, this feature may well be entirely useless in practise. A more recent paper by this group (abstract here, subscription required for full paper), succeeds in using DNA tiles to carry out some elementary calculations, but highlights the difficulties caused by the significant error rate in the elementary operations. Secondly, this offers, in principle, a very effective way of designing and executing very complicated and rich structures that combine design with, in some cases, aperiodicity. In the physical realisation here, the starting conditions are specified by the sequence of a “seed” strand of DNA, while the rule is embodied in the design of the sticky patches on the tiles, itself specified by the sequence of the DNA from which they are made. Simple modifications of the seed strand sequence and the rule implicit in the tile design could result in a wide and rich design space of resulting “algorithmic crystals”.
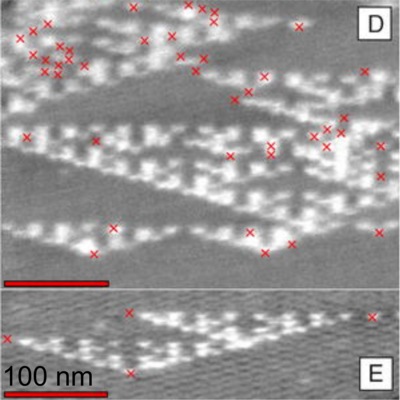
A physical realisation of a cellular automaton executed using self-assembling DNA tiles. Red crosses indicate propagation errors, which intiatiate or terminate the characteristic Sierpinski triangle patterns. From Rothemund et al, PLOS Biology 2 2041 (2004), copyright the authors, reproduced under a CREATIVE COMMONS ATTRIBUTION LICENSE