It currently costs about a million dollars to sequence an individual human genome. One can expect incremental improvements in current technology to drop this price to around $100,000, but the need that current methods have to amplify the DNA will make it difficult for this price to drop further. So, to meet a widely publicised target of a $1000 genome a fundamentally different technology is needed. One very promising approach uses the idea of threading a single DNA molecule through a nanopore in a membrane, and identifiying each base by changes in the ion current flowing through the pore. I wrote about this a couple of years ago, and a talk I heard yesterday from one of the leaders in the field prompts me to give an update.
The original idea for this came from David Deamer and Dan Branton, who filed a patent for the general scheme in 1998. Hagan Bayley, from Oxford, whose talk I heard yesterday, has been collaborating with Reza Ghadiri from Scripps, to implement this scheme using a naturally occuring pore forming protein, alpha-hemolysin, as the reader.
The key issues are the need to get resolution at a single base level, and the correct identification of the bases. They get extra selectivity by a combination of modification of the pore by genetic engineering, and insertion into the pore of small ring molecules – cyclodextrins. At the moment speed of reading is a problem – when the molecules are pulled through by an electric field they tend to go a little too fast. But, in an alternative scheme in which bases are chopped off the chain one by one and dropped into the pore sequentially, they are able to identify individual bases reliably.
Given that the human genome has about 6 million bases, they estimate that at 1 millisecond reading time per base they’ll need to use 1000 pores in parallel to sequence a genome in under a day (taking into account the need for a certain amount of redundancy for error correction). To prepare the way for commercialisation of this technology, they have a start-up company – Oxford NanoLabs – which is working on making a miniaturised and rugged device, about the size of a palm-top computer, to do this kind of analysis.
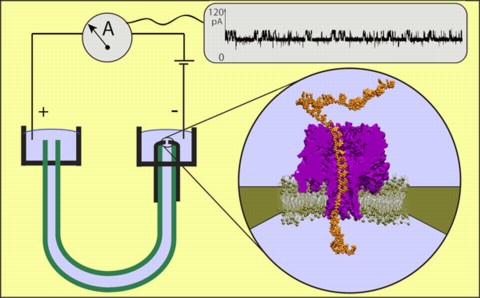
Schematic of a DNA reader using the pore forming protein alpha-hemolysin. As the molecule is pulled through the pore, the ionic conduction through the pore varies, giving a readout of the sequence of bases. From the website of the Theoretical and Computational Biophysics group at the University of Illinois at Urbana-Champaign.
I can see it now, reminiscent of the ‘Airport’ scene from Fifth Element. The Human Identification Police stopping anyone who does not look ‘homo sapien’ enough and doing the instant genome screen.
A para phrase of the first encounter comment by one of the Nano Bio members of the crew.
The reality is however, this is yet another advance, which in the first instance will give more opportunity for those within the ‘Structured Laboratory’ community, that is those working to a specific agenda, and in the long term, opening the field of discovery to those who are interested in the knowledge that comes with discovery and rediscovery and the further dissemination of it.
The Revolution/Evolution continues.
Would reducing the electric field strength slow the velocity of the DNA through the cyclodextrin?
Sir,…
If you havent heard already, bionanomatrix has recently been awarded a project for a $100 genome…
http://nanotechnology.net/index.aspx?ID=86256
Martin, Hagan illustrated his talk with a still from the film Gattaca, so those science fiction visions really do have an influence on how people think about their science.
Phillip, I suspect that the problem is that as you increase the electric field, the DNA doesn’t go anywhere at all, and then you reach some critical field it all rushes out at once, as it were. I think their approach is to modify the frictional properties of the inside of the pore, both by genetically engineering the protein and by changing the ring-like “washer” molecule that they put in the pore.
Dibyadeep, good luck to bionanomatrix but let’s see if we can get to $1000 before worrying about $100!
Richard – Agreed, I am reminded of a tour I took of Pinewood studios many years ago and they were working on one of the ‘Bond’ films. Along side the fictional ‘Q’ were a group of technical people making sure everything not only appeared to work, but actually worked. One of Albert R. Broccoli’s cardinal rules was that ‘it has to be real’.
Many things that are written about in fiction are either related to fact or speak at least in a cursory way to ‘Works in Progress’
One of the crew is working on the Mathematical equations involved in cloaking. Someone suggested he consult with the Romulans until he demonstrated that it is a legitimate area being studied.
I would suggest that much that is being invented today has been thought of or dreamed about for years. The age we live in today, the developing collaborative nature of this very venue make the dreams much closer or faster to reality.
Very cool work, I can’t wait to have more than just a 256-bit Hex number ID 🙂
So an electric field pulls the DNA strand through to read each individual codone?
I’m regretting my physics textbook is mostly still a paperweight. I’d think that it would be possible to functionalize the DNA in solution-phase to slow the needle-threading enough to be readable, but I’ll come back to this thread when/if I have a specific chemistry in mind.
I saw a presentation by Bayley a couple of years ago. He said then that one possible use was to help in reintegrating the data from shotgun reading techniques. Even though he couldn’t get an accurate read of every base, he could get a ‘blurred’ picture over the whole length of the DNA. So his data could resolve positioning ambiguities when reassembling the shotgun sequences. I don’t know whether that’s been done yet.
As regards slowing the molecule down, I wonder if it’s possible to attach a sphere to one end – perhaps using a specific linker – and then use an optical trap to slow it down.
Cheers, Dave
There was an interesting paper in PNAS recently about characterizing the T4 phage packaging motor. It gives an example of how optical traps can be used to control DNA.
Potentially even more interesting, though, is what might be done given what they discovered. It seems that it might be possible to develop a system that uses the T4 motor itself to control the passage of a DNA molecule.
It might then be possible to engineer the motor to bind to alpha-hemolysin to make a DNA reader. I suppose it’s conceivable that there may be some property of the motor itself that is sensitive to the individual bases in the DNA and might be made externally observable.
I put a URL in my last comment, but it’s disappeared? Hopefully it can be retrieved
Dave, I’m sorry, there’s no trace of the URL. Did you put it in as a link or just as an address?
But I think this is the article you referred to…
http://www.pnas.org/cgi/content/abstract/104/43/16868
Very interesting indeed, for the reasons you say.