Gene therapy holds out the promise of correcting a number of diseases whose origin lies in the deficiency of a particular gene – given our growing knowledge of the human genome, and our ability to synthesise arbitrary sequences of DNA, one might think that the introduction of new genetic material into cells to remedy the effects of abnormal genes would be straightforward. This isn’t so. DNA is a relatively delicate molecule, and organisms have evolved efficient mechanisms for finding and eliminating foreign DNA. Viruses, on the other hand, whose entire modus operandi is to introduce foreign nucleic acids into cells, have evolved effective ways of packaging their payloads of DNA or RNA into cells. One approach to gene therapy co-opts viruses to deliver the new genetic material, though this sometimes has unpredicted and undesirable side-effects. So an effective, non-viral method of wrapping up DNA, introducing it into target cells and releasing it would be very desirable. My colleagues at Sheffield University, led by Beppe Battaglia, have recently demonstrated an effective and elegant way of introducing DNA into cells, in work recently reported in the journal Advanced Materials (subscription required for full paper).
The technique is based on the use of polymersomes, which I’ve described here before. Polymersomes are bags formed when detergent-like polymer molecules self-assemble to form a membrane which folds round on itself to form a closed surface. They are analogous to the cell membranes of biology, which are formed from soap-like molecules called phospholipids, and the liposomes that can be made in the laboratory from the same materials. Liposomes are used to wrap up and deliver molecules in some commercial applications already, including some drug delivery systems and in some expensive cosmetics. They’ve also been used in the laboratory to deliver DNA into cells, though they aren’t ideal for this purpose, as they aren’t very robust. Polymersomes allow one a great deal more flexibility in designing polymersomes with the properties one needs, and this flexibility is exploited to the full in Battaglia’s experiments.
To make a polymersome, one needs a block copolymer – a polymer with two or three chemically distinct sections joined together. One of these blocks needs to be hydrophobic, and one needs to be hydrophilic. The block copolymers used here, developed and synthesised in the group of Sheffield chemist Steve Armes, have two very nice features. The hydrophilic section is composed of poly(2-(methacryloyloxy)ethyl phosphorylcholine) – this is a synthetic polymer that presents the same chemistry to the adjoining solution as a naturally occurring phospholipid in a cell membrane. This means that polymersomes made from this material are able to circulate undetected within the body for longer than other water soluble polymers. The hydrophobic block is poly(2-(diisopropylamino)ethyl methacrylate). This is a weak base, so it has the property that its state of ionisation depends on the acidity of the solution. In a basic solution, it is un-ionized, and in this state it is strongly hydrophobic, while in an acidic solution it becomes charged, and in this state it is much more soluble in water. This means that polymersomes made from this material will be stable in neutral or basic conditions, but will fall apart in acid. Conversely, if one has the polymers in an acidic solution, together with the DNA one wants to deliver, and then neutralises the solution, polymersomes will spontaneously form, encapsulating the DNA.
The way these polymersomes work to introduce DNA into cells is sketched in the diagram below. On encountering a cell, the polymersome triggers the process of endocytosis, whereby the cell engulfs the polymersome in a little piece of cell membrane that is pinched off inside the cell. It turns out that the solution inside these endosomes is significantly more acidic than the surroundings, and this triggers the polymersome to fall apart, releasing its DNA. This, in turn, generates an osmotic pressure sufficient to burst open the endosome, releasing the DNA into the cell interior, where it is free to make its way to the nucleus.
The test of the theory is to see whether one can introduce a section of DNA into a cell and then demonstrate how effectively the corresponding gene is expressed. The DNA used in these experiments was the gene that codes for a protein that fluoresces – the famous green fluorescent protein, GFP, originally obtained from certain jelly-fish – making it easy to detect whether the protein coded for by the introduced gene has actually been made. In experiments using cultured human skin cells, the fraction of cells in which the new gene was introduced was very high, while few toxic effects were observed, in contrast to a control experiment using an existing, commercially available gene delivery system, which was both less effective at introducing genes and actually killed a significant fraction of the cells.
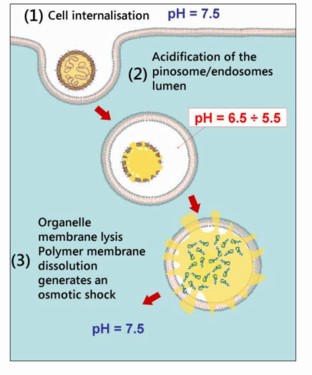
A switchable polymersome as a vehicle for gene delivery. Beppe Battaglia, University of Sheffield.